Cross-categorization of legal concepts across boundaries of legal systems: in consideration of inferential links by Fumiko Kano Glückstad, Tue Herlau, Mikkel N. Schmidt, Morten Mørup.
Abstract:
This work contrasts Giovanni Sartor’s view of inferential semantics of legal concepts (Sartor in Artif Intell Law 17:217–251, 2009) with a probabilistic model of theory formation (Kemp et al. in Cognition 114:165–196, 2010). The work further explores possibilities of implementing Kemp’s probabilistic model of theory formation in the context of mapping legal concepts between two individual legal systems. For implementing the legal concept mapping, we propose a cross-categorization approach that combines three mathematical models: the Bayesian Model of Generalization (BMG; Tenenbaum and Griffiths in Behav Brain Sci 4:629–640, 2001), the probabilistic model of theory formation, i.e., the Infinite Relational Model (IRM) first introduced by Kemp et al. (The twenty-first national conference on artificial intelligence, 2006, Cognition 114:165–196, 2010) and its extended model, i.e., the normal-IRM (n-IRM) proposed by Herlau et al. (IEEE International Workshop on Machine Learning for Signal Processing, 2012). We apply our cross-categorization approach to datasets where legal concepts related to educational systems are respectively defined by the Japanese- and the Danish authorities according to the International Standard Classification of Education. The main contribution of this work is the proposal of a conceptual framework of the cross-categorization approach that, inspired by Sartor (Artif Intell Law 17:217–251, 2009), attempts to explain reasoner’s inferential mechanisms.
From the introduction:
An ontology is traditionally considered as a means for standardizing knowledge represented by different parties involved in communications (Gruber 1992; Masolo et al. 2003; Declerck et al. 2010). Kemp et al. (2010) also points out that some scholars (Block 1986; Field 1977; Quilian 1968) have argued the importance of knowledge structuring, i.e., ontologies, where concepts are organized into systems of relations and the meaning of a concept partly depends on its relationships to other concepts. However, real human to human communication cannot be absolutely characterized by such standardized representations of knowledge. In Kemp et al. (2010), two challenging issues are raised against such idea of systems of concepts. First, as Fodor and Lepore (1992) originally pointed out, it is beyond comprehension that the meaning of any concept can be defined within a standardized single conceptual system. It is unrealistic that two individuals with different beliefs have common concepts. This issue has also been discussed in semiotics (Peirce 2010; Durst-Andersen 2011) and in cognitive pragmatics (Sperber and Wilson 1986). For example, Sperber and Wilson (1986) discuss how mental representations are constructed diversely under different environmental and cognitive conditions. A second point which Kemp et al. (2010) specifically address in their framework is the concept acquisition problem. According to Kemp et al. (2010; see also: Hempel (1985), Woodfield (1987)):
if the meaning of each concept depends on its role within a system of concepts, it is difficult to see how a learner might break into the system and acquire the concepts that it contains. (Kemp et al. 2010)
Interestingly, the similar issue is also discussed by legal information scientists. Sartor (2009) argues that:
legal concepts are typically encountered in the context of legal norms, and the issue of determining their content cannot be separated from the issue of identifying and interpreting the norms in which they occur, and of using such norms in legal inference. (Sartor 2009)
This argument implies that if two individuals who are respectively belonging to two different societies having different legal systems, they might interpret a legal term differently, since the norms in which the two individuals belong are not identical. The argument also implies that these two individuals must have difficulties in learning a concept contained in the other party’s legal system without interpreting the norms in which the concept occurs.
These arguments motivate us to contrast the theoretical work presented by Sartor (2009) with the probabilistic model of theory formation by Kemp et al. (2010) in the context of mapping legal concepts between two individual legal systems. Although Sartor’s view addresses the inferential mechanisms within a single legal system, we argue that his view is applicable in a situation where a concept learner (reasoner) is, based on the norms belonging to his or her own legal system, going to interpret and adapt a new concept introduced from another legal system. In Sartor (2009), the meaning of a legal term results from the set of inferential links. The inferential links are defined based on the theory of Ross (1957) as:
- the links stating what conditions determine the qualification Q (Q-conditioning links), and
- the links connecting further properties to possession of the qualification Q (Q-conditioned links.) (Sartor 2009)
These definitions can be seen as causes and effects in Kemp et al. (2010). If a reasoner is learning a new legal concept in his or her own legal system, the reasoner is supposed to seek causes and effects identified in the new concept that are common to the concepts which the reasoner already knows. This way, common-causes and common-effects existing within a concept system, i.e., underlying relationships among domain concepts, are identified by a reasoner. The probabilistic model in Kemp et al. (2010) is supposed to learn these underlying relationships among domain concepts and identify a system of legal concepts from a view where a reasoner acquires new concepts in contrast to the concepts already known by the reasoner.
Pardon the long quote but the paper is pay-per-view.
I haven’t started to run down all the references but this is an interesting piece of work.
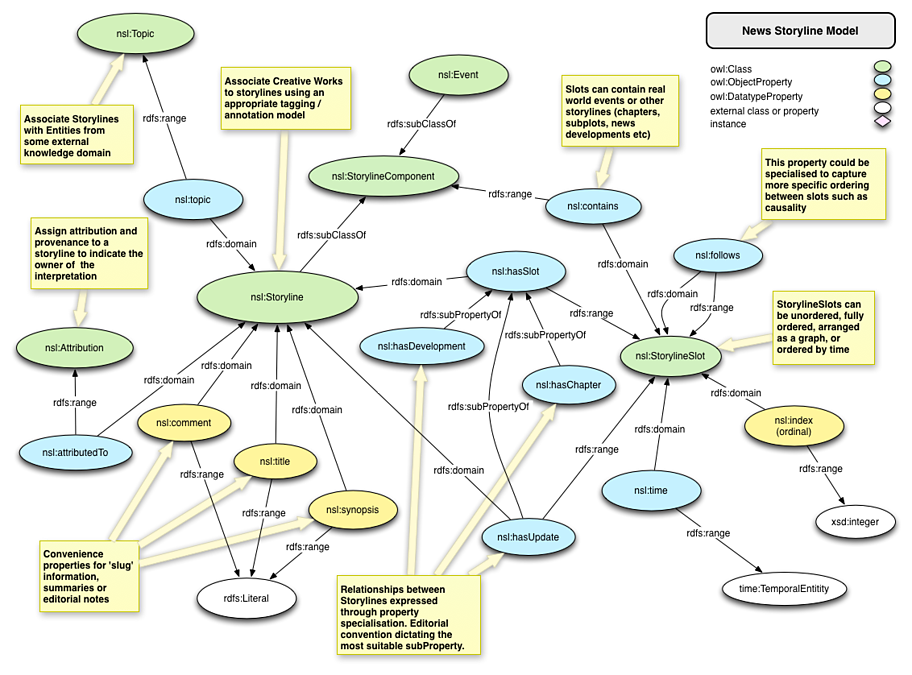
I was most impressed by the partial echoing of the topic map paradigm that: “meaning of each concept depends on its role within a system of concepts….”
True, a topic map can capture only “surface” facts and relationships between those facts but that merits a comment on a topic map instance and not topic maps in general.
Noting that you also shouldn’t pay for more topic map than you need. If all you need is a flat mapping between DHS and say the CIA, then doing nor more than mapping terms is sufficient. If you need a maintainable and robust mapping, different techniques would be called for. Both results would be topic maps, but one of them would be far more useful.
One of the principal sources relied upon by the authors’ is: The Nature of Legal Concepts: Inferential Nodes or Ontological Categories? by Giovanni Sartor.
I don’t see any difficulty with Sartor’s rules of inference, any more than saying if a topic has X property (occurrence in TMDM speak), then of necessity it must have property E, F, and G.
Where I would urge caution is with the notion that properties of a legal concept spring from a legal text alone. Or even from a legal ontology. In part because two people in the same legal system can read the same legal text and/or use the same legal ontology and expect to see different properties for a legal concept.
Consider the text of Paradise Lost by John Milton. If Stanley Fish, a noted Milton scholar, were to assign properties to the concepts in Book 1, his list of properties would be quite different from my list of properties. Same words, same text, but very different property lists.
To refine what I said about the topic map paradigm a bit earlier, it should read: “meaning of each concept depends on its role within a system of concepts [and the view of its hearer/reader]….”
The hearer/reader being the paramount consideration. Without a hearer/reader, there is no concept or system of concepts or properties of either one for comparison.
When topics are merged, there is a collecting of properties, some of which you may recognize and some of which I may recognize, as identifying some concept or subject.
No guarantees but better than repeating your term for a concept over and over again, each time in a louder voice.