Activities at the Royal Society of Chemistry to gather, extract and analyze big datasets in chemistry by Antony Williams.
If you are looking for a quick summary of efforts to combine existing knowledge resources in chemistry, you can do far worse than Antony’s 118 slides on the subject (2015).
I want to call special attention to Slide 107 in his slide deck:
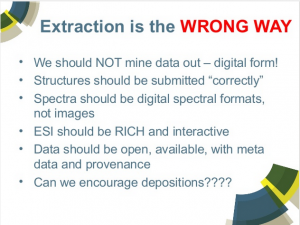
True enough, extraction is problematic, expensive, inaccurate, etc., all the things Antony describes. And I would strongly second all of what he implies is the better practice.
However, extraction isn’t just a necessity for today or for a few years, extraction is going to be necessary so long as we keep records about chemistry or any other subject.
Think about all the legacy materials on chemistry that exist in hard copy format just for the past two centuries. To say nothing of all of still older materials. It is more than unfortunate to abandon all that information simply because “modern” digital formats are easier to manipulate.
That was’t what Antony meant to imply but even after all materials have been extracted and exist in some form of digital format, that doesn’t mean the era of “extraction” will have ended.
You may not remember when atomic chemistry used “punch cards” to record isotopes:

An isotope file on punched cards. George M. Murphy J. Chem. Educ., 1947, 24 (11), p 556 DOI: 10.1021/ed024p556 Publication Date: November 1947.
Today we would represent that record in…NoSQL?
Are you confident that in another sixty-eight (68) years we will still be using NoSQL?
We have to choose from the choices available to us today, but we should not deceive ourselves into thinking our solution will be seen as the “best” solution in the future. New data will be discovered, new processes invented, new requirements will emerge, all of which will be clamoring for a “new” solution.
Extraction will persist as long as we keep recording information in the face of changing formats and requirements. We can improve that process but I don’t think we will ever completely avoid it.

