SCADA Researcher Drops Zero-Day, ICS-CERT Issues Advisory by Kelly Jackson Higgins. (A patch has been released: Advisory (ICSA-14-016-01))
From the post:
A well-known and prolific ICS/SCADA vulnerability researcher here today revealed a zero-day flaw in a Web server-based system used for monitoring, controlling, and viewing devices and systems in process control environments.
Luigi Auriemma, CEO of Malta-based zero-day vulnerability provider and penetration testing firm ReVuln, showed a proof-of-concept for executing a buffer overflow attack on Ecava’s IntegraXor software, which is used in human machine interfaces (HMIs) for SCADA systems.
The ICS-CERT responded later in the day with a security alert on the zero-day vulnerability, and requested that Ecava confirm the bug and provide mitigation. Ecava as of this posting had not responded publicly, nor had it responded to an email inquiry by Dark Reading.
The IntegraXor line is used in process control environments in 38 countries, mainly in the U.K., U.S., Australia, Poland, Canada, and Estonia, according to ICS-CERT.
How bad is this?
First, zero-day is a media hype term. Essentially means the vendor finds out about the flaw as the same time it is made public.
Second, what are SCADA systems?
The summary in Wikipedia reads:
SCADA (supervisory control and data acquisition) is a type of industrial control system (ICS). Industrial control systems are computer-controlled systems that monitor and control industrial processes that exist in the physical world. SCADA systems historically distinguish themselves from other ICS systems by being large-scale processes that can include multiple sites, and large distances. [footnote [1] omitted] These processes include industrial, infrastructure, and facility-based processes, as described below:
- Industrial processes include those of manufacturing, production, power generation, fabrication, and refining, and may run in continuous, batch, repetitive, or discrete modes.
- Infrastructure processes may be public or private, and include water treatment and distribution, wastewater collection and treatment, oil and gas pipelines, electrical power transmission and distribution, wind farms, civil defense siren systems, and large communication systems.
- Facility processes occur both in public facilities and private ones, including buildings, airports, ships, and space stations. They monitor and control heating, ventilation, and air conditioning systems (HVAC), access, and energy consumption.
The geographic range of vulnerable systems was specified in the original CERT alert:
IntegraXor is a suite of tools used to create and run a Web-based human-machine interface for a SCADA system. IntegraXor is currently used in several areas of process control in 38 countries with the largest installation based in the United Kingdom, United States, Australia, Poland, Canada, and Estonia. (emphasis added)
From a follow up CERT alert, VULNERABILITY DETAILS:
EXPLOITABILITY
This vulnerability could be exploited remotely.
EXISTENCE OF EXPLOIT
Exploits that target this vulnerability are publicly available.
DIFFICULTY
An attacker with a low skill would be able to exploit this vulnerability.
The top six geographic locations:
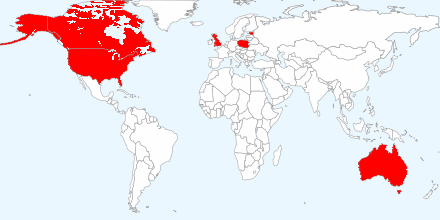
have vulnerabilities in: industrial processes, manufacturing, production, power generation, fabrication, and refining; infrastructure processes, water treatment and distribution, wastewater collection and treatment, oil and gas pipelines, electrical power transmission and distribution, wind farms, civil defense siren systems, and large communication systems; facility processes, buildings, airports, ships, and space stations.
due to buffer overflows.
According to Wikipedia, the earliest documentation on buffer overflows dates from 1972 and the first hostile exploit of a buffer overflow was in 1988 (the Morris worm).
A quick search on integraxor “buffer overflow” returned 1,850 “hits,” most of them duplicates of the original news or opinions about the infrastructures at risk.
But there was one “buffer overflow” POC with Integraxor in 2010. But you had to sift through a number of “hits” to find it.
Experts already know where to find buffer overflow opportunities but it doesn’t appear that level of expertise is widely shared.
It appears to be time consuming, but not difficult, to identify customers of particular vendors to approach with security services in the event of exploits.
For a security flaw story, what would you want to know that can’t be learned at Exploit Database?
What facts, other data sources, organization of that information, etc.?
Thinking topic maps as “in addition to” rather than “an alternative for” some existing information store, will be easier to sell.