I encountered a wonderful example of “overlap” in the markup sense today while reading about resolving conflicts in constructing a comprehensive tree of life.
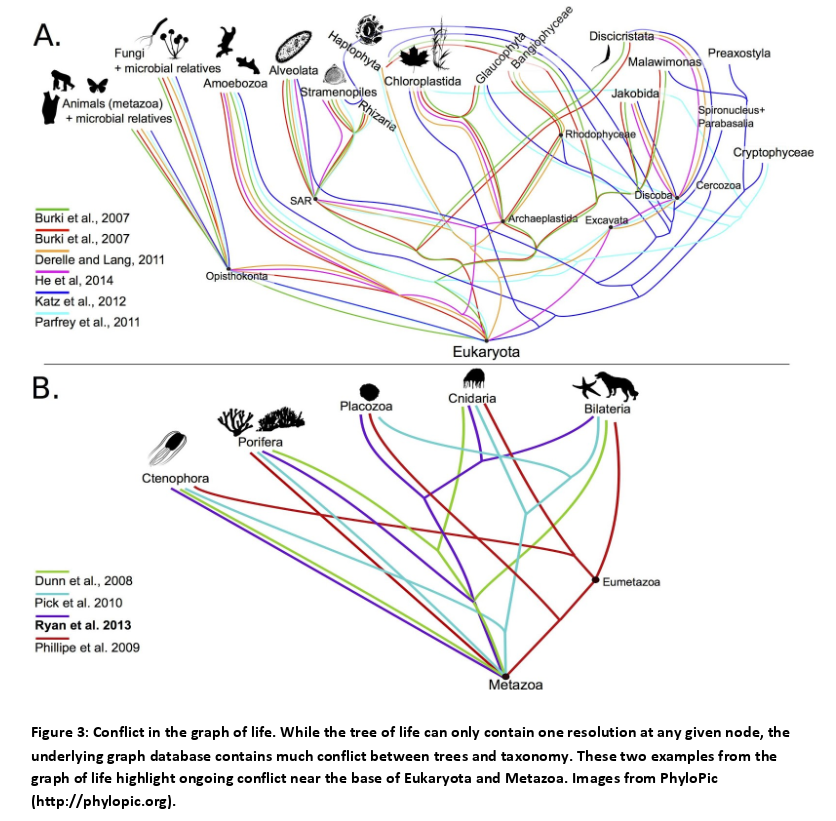
The authors use a graph database which allows them to study various hypotheses on the resolutions of conflicts.
Their graph database, opentree-treemachine, is available on GitHub, https://github.com/OpenTreeOfLife/treemachine, as is the source to all the project’s software, https://github.com/OpenTreeOfLife.
There’s a thought for Balisage 2015. Is the processing of overlapping markup a question of storing documents with overlapping markup in graph databases and then streaming the non-overlapping results of a query to an XML processor?
And visualizing overlapping results or alternative resolutions to overlapping results via a graph database.
The question of which overlapping syntax to use becoming a matter of convenience and the amount of information captured, as opposed to attempts to fashion syntax that cheats XML processors and/or developing new means for processing XML.
Perhaps graph databases can make overlapping markup in documents the default case just as overlap is the default case in documents (single tree documents being rare outliers).
Remind me to send a note to Michael Sperberg-McQueen and friends about this idea.
BTW, the details of the article that lead me down this path:
Synthesis of phylogeny and taxonomy into a comprehensive tree of life by Steven A. Smith, et al.
Abstract:
Reconstructing the phylogenetic relationships that unite all biological lineages (the tree of life) is a grand challenge of biology. However, the paucity of readily available homologous character data across disparately related lineages renders direct phylogenetic inference currently untenable. Our best recourse towards realizing the tree of life is therefore the synthesis of existing collective phylogenetic knowledge available from the wealth of published primary phylogenetic hypotheses, together with taxonomic hierarchy information for unsampled taxa. We combined phylogenetic and taxonomic data to produce a draft tree of life—the Open Tree of Life—containing 2.3 million tips. Realization of this draft tree required the assembly of two resources that should prove valuable to the community: 1) a novel comprehensive global reference taxonomy, and 2) a database of published phylogenetic trees mapped to this common taxonomy. Our open source framework facilitates community comment and contribution, enabling a continuously updatable tree when new phylogenetic and taxonomic data become digitally available. While data coverage and phylogenetic conflict across the Open Tree of Life illuminates significant gaps in both the underlying data available for phylogenetic reconstruction and the publication of trees as digital objects, the tree provides a compelling starting point from which we can continue to improve through community contributions. Having a comprehensive tree of life will fuel fundamental research on the nature of biological diversity, ultimately providing up-to-date phylogenies for downstream applications in comparative biology, ecology, conservation biology, climate change studies, agriculture, and genomics.
A project with a great deal of significance beyond my interest in overlap in markup documents. Highly recommended reading. The resolution of conflicts in trees here involves an evaluation of data, much as you would for merging in a topic map.
Unlike the authors, I see no difficulty in super trees being rich enough with the underlying data to permit direct use of trees for resolution of conflicts. But you would have to design the trees from the start with those capabilities or have topic map like merging capabilities so you are not limited by early and necessarily preliminary data design decisions.
Enjoy!
I first saw this in a tweet by Ross Mounce.