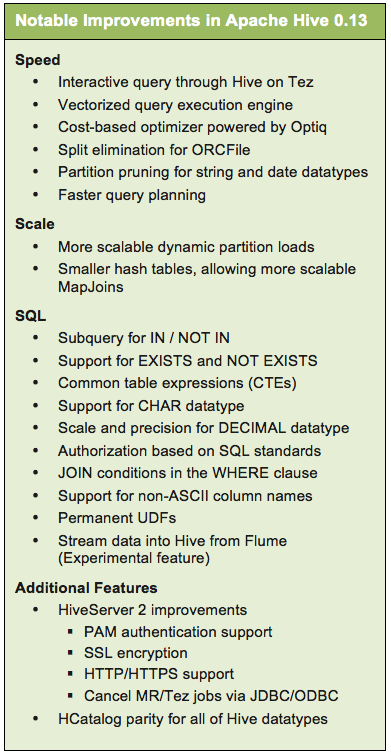
Depending on your community, when you hear “Hive,” you think “Apache Hive:”
The Apache Hive ™ data warehouse software facilitates querying and managing large datasets residing in distributed storage. Hive provides a mechanism to project structure onto this data and query the data using a SQL-like language called HiveQL. At the same time this language also allows traditional map/reduce programmers to plug in their custom mappers and reducers when it is inconvenient or inefficient to express this logic in HiveQL.
But, there is another “Hive,” which handles large datasets:
High-performance Integrated Virtual Environment (HIVE) is a specialized platform being developed/implemented by Dr. Simonyan’s group at FDA and Dr. Mazumder’s group at GWU where the storage library and computational powerhouse are linked seamlessly. This environment provides web access for authorized users to deposit, retrieve, annotate and compute on HTS data and analyze the outcomes using web-interface visual environments appropriately built in collaboration with research scientists and regulatory personnel.
I ran across this potential source of confusion earlier today and haven’t run it completely to ground but wanted to share some of what I have found so far.
Inside the HIVE, the FDA’s Multi-Omics Compute Architecture by Aaron Krol.
From the post:
“HIVE is not just a conventional virtual cloud environment,” says Simonyan. “It’s a different system that virtualizes the services.” Most cloud systems store data on multiple servers or compute units until users want to run a specific application. At that point, the relevant data is moved to a server that acts as a node for that computation. By contrast, HIVE recognizes which storage nodes contain data selected for analysis, then transfers executable code to those nodes, a relatively small task that allows computation to be performed wherever the data is stored. “We make the computations on exactly the machines where the data is,” says Simonyan. “So we’re not moving the data to the computational unit, we are moving computation to the data.”
When working with very large packets of data, cloud computing environments can sometimes spend more time on data transfer than on running code, making this “virtualized services” model much more efficient. To function, however, it relies on granular and readily-accessed metadata, so that searching for and collecting together relevant data doesn’t consume large quantities of compute time.
HIVE’s solution is the honeycomb data model, which stores raw NGS data and metadata together on the same network. The metadata — information like the sample, experiment, and run conditions that produced a set of NGS reads — is stored in its own tables that can be extended with as many values as users need to record. “The honeycomb data model allows you to put the entire database schema, regardless of how complex it is, into a single table,” says Simonyan. The metadata can then be searched through an object-oriented API that treats all data, regardless of type, the same way when executing search queries. The aim of the honeycomb model is to make it easy for users to add new data types and metadata fields, without compromising search and retrieval.
Popular consumption piece so next you may want to visit the HIVE site proper.
From the webpage:
HIVE is a cloud-based environment optimized for the storage and analysis of extra-large data, like Next Generation Sequencing data, Mass Spectroscopy files, Confocal Microscopy Images and others.
HIVE uses a variety of advanced scientific and computational visualization graphics, to get the MOST from your HIVE experience you must use a supported browser. These include Internet Explore 8.0 or higher (Internet Explorer 9.0 is recommended), Google Chrome, Mozilla Firefox and Safari.
A few exemplary analytical outputs are displayed below for your enjoyment. But before you can take advantage of all that HIVE has to offer and create these objects for yourself, you’ll need to register.
With A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE) by Tsung-Jung Wu, et al., you are starting to approach the computational issues of interest for data integration.
From the article:
The forementioned cooperation is difficult because genomics data are large, varied, heterogeneous and widely distributed. Extracting and converting these data into relevant information and comparing results across studies have become an impediment for personalized genomics (11). Additionally, because of the various computational bottlenecks associated with the size and complexity of NGS data, there is an urgent need in the industry for methods to store, analyze, compute and curate genomics data. There is also a need to integrate analysis results from large projects and individual publications with small-scale studies, so that one can compare and contrast results from various studies to evaluate claims about biomarkers.
See also: High-performance Integrated Virtual Environment (Wikipedia) for more leads to the literature.
Heterogeneous data is still at large and people are building solutions. Rather than either/or, what do you think topic maps could bring as a value-add to this project?
I first saw this in a tweet by ChemConnector.